Cancer cell metastasis. Stem cell differentiation. Atherosclerosis. All are strong mechanotransduction-related physiological and pathophysiological events. Just how do cells transduce mechanical force into biochemical signals?

“Cells are sensitive to mechanical forces outside the cell membrane,” says Sangyoon Han, who joined the Department of Biomedical Engineering at Michigan Tech as an assistant professor last fall. At their basal surface, however, cells are interfacing with something called the extracellular matrix (ECM), which supports the cell not only chemically but also mechanically.”
“Over the past 20 years, it has been revealed that the rigidity of the extracellular matrix can greatly influence the physiology and pathology of cells and tissues, including differentiation, survival, proliferation, altered drug response, and tumor progression,” adds Han. “In the case of a tumor, an increase in tissue stiffness—without any changes in genetic information and chemical environment—can cause tumor progression. There is also an evidence showing that cancer-targeting drugs do not work when cancer cells are highly contractile in a very tensed environment,” he says.
To investigate this, Han and his team established experimental and computational frameworks for force measurement and adhesion dynamics quantification. “We apply these frameworks, with cutting-edge computer vision techniques, on live-cell microscope images to find out the fundamental mechanisms underlying mechanosensation in normal cells, as well as the biomechanical signature in diseased cells whose signaling has gone awry.”
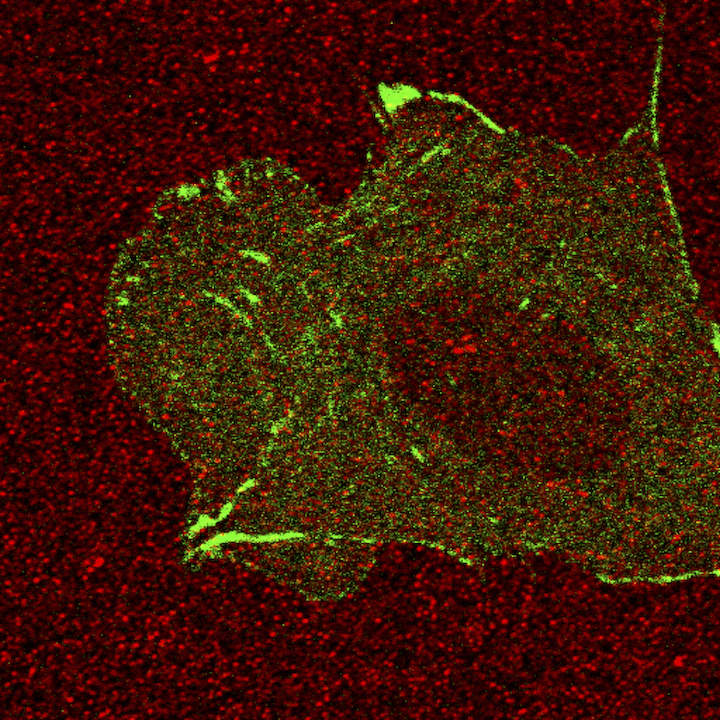
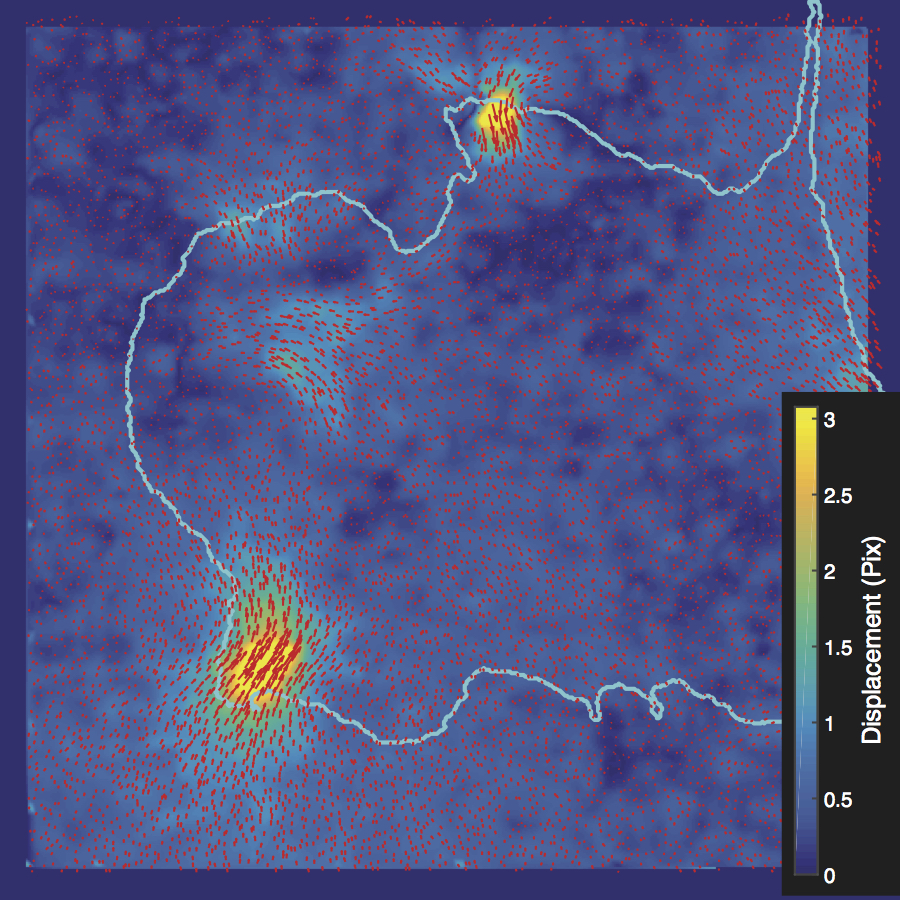
Han measures the force a cell transmits to the environment with traction force microscopy. “The force sensor, referred to as a focal adhesion, consists of a special receptor across the membrane and over 100 cytoskeletal adaptor proteins. These focal adhesion proteins have redundant and diverse roles in signaling and structural development of the adhesion,” he explains.
Using high-resolution imaging of living cells on a soft substrate, Han captures gel deformation and force-sensing protein trajectories at the same time. Han’s novel force-reconstruction software converts the measured gel deformation into a force map over a cell footprint. Using time-series data extracted from the image data, he monitors feedback between the cellular structure and its mechanical forces.
Han shares his Matlab-based, open-source software with the mechanobiology community. In his Mechanobiology Lab at Michigan Tech, Han is also building a physical device using bioMEMS for active force application to cells and tissue. “I firmly believe that engineers can make significant contributions to not only the biomedical industry, but also fundamental biological science.”
Before coming to Michigan Tech, Han was a postdoctoral researcher at the Harvard Medical School Lab of Computational Cell Biology, as well as the University of Texas Southwestern Medical Center. He earned a PhD in Mechanical Engineering at University of Washington in the area of cell mechanics, multiphysics modeling, and bioMEMS, and BS and MS in Mechanical Engineering at Seoul National University.
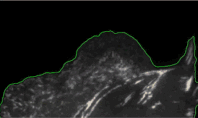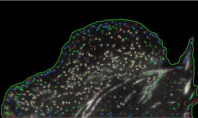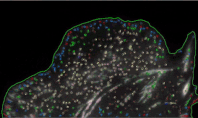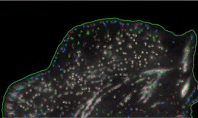
Red spots are the fluorescent beads coated on top of the gel, which we use to quantify the deformation of the gel. Green signal is the paxillin, one of the focal adhesion proteins of a Chinese Hamster ovary cell.Sangyoon Han, assistant professor of biomedical engineering at Michigan Technological University